By Dana Christensen
Nov 15, 2016
The DevOps movement is focused on leading transformational change and driving innovation. At the recent DevOps Enterprise Summit in San Francisco, many of the leaders in the field spoke of driving change through a culture focused on collaboration, community, co-creation, curiosity, continual learning, designing for joy, and meaningful work. I have been impressed with the passion and conviction that leaders in the DevOps movement speak about and emphasize these key principles.. Through determination and focus, living out these key values, and recognizing that everyone has a role to play—we will be able to truly unlock the power of technology to address the many complex global challenges we face today.
An excellent example of leveraging DevOps and technology to address complex global challenges is found in the field of Genomics Research. Through focus on the values spoken about by DevOps thought leaders and innovation at the speed of community—Science, Universities, Government, and Business, powered by advances in IT, are able to join forces to develop and evolve techniques that allow for the reading of the source code of biology—something that is incredibly complex and in parts extremely optimized. Through this important work, we are just beginning to unlock the secrets and miraculous mysteries to life on earth as we know it.
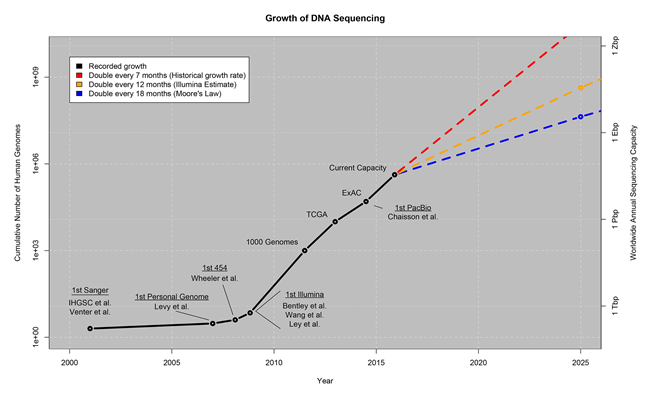
The field of genomics puts the “Big” in Big Data. In short, it is projected that by 2025 genomics will produce about 1 zetta-bases per year (that is roughly the same amount in zeta-bytes) – following a trend of doubling in sequencing capacity every 12 months. Genomics presents some of the most demanding computational requirements that we will face in the coming decade. These challenges will only be met through the power of community—where generative collaboration, co-creation, trust and sharing can can create an environment conducive to creating solutions that will unleash the power of genomics sequencing.
The PLOS article Big Data: Astromical or Genomical? provides an interesting overview of the challenges faced when managing this amount of data.
As discussed in the PLOS article, there are four components that comprise the “life cycle” of the Genomics dataset: Acquisition, Storage, Data Distribution, and Analysis. Each of these domains contains it’s own set of challenges for the community.
In the area of data acquisition, in order to sustain the explosive growth in genomic data sequencing, it is critical to advance the development and application of technologies that reduce cost, increase throughput, and minimize human errors—all of which can only be accomplished at scale with automation. This is where StackStorm fits into the equation.
I recently had the opportunity to connect with members of the StackStorm community from SciLifeLab, a national center for molecular bioscience, hosted by Uppsala University, Stockholm University, KTH and the Karolinska Institute. The team there is responsible for the development and operations of SNP&SEQ (http://www.sequencing.se), a technology platform that provides sequencing and genotyping services for researchers, primarily within Sweden, but also some from abroad. Together with a couple of other sequencing platforms in Uppsala and Stockholm, they comprise the National Genomics Infrastructure, NGI, which is the largest technology platform within SciLifeLab, and one of the biggest sequencing centers in Europe (https://www.scilifelab.se/platforms/ngi/).
This year SNP&SEQ is expected to produce roughly 500 TB of data – following the industry trend of doubling capacity every 12 months. At SciLifeLab, the team’s answer to the challenge of scaling, while lowering costs, increasing throughput, and minimizing human errors is the Arteria Project (https://arteria-project.github.io/).
The team has leveraged the power of StackStorm as a hub to automate and streamline their complex sequencing workflows. StackStorm has played an instrumental role in allowing the facility to continue to scale with a relatively small team managing sequence operations. By leveraging StackStorm for automation, the team has been able to focus on the specifics of their “business case” rather than building up their own systems and interfaces.
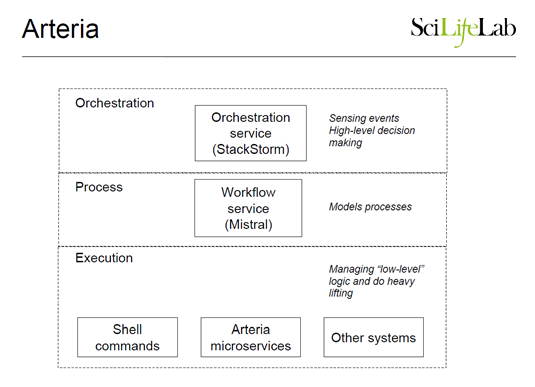
StackStorm is being used primarily to drive their main processing and quality control pipeline. The implementation consists of microservices running on a local compute farm, and a master node running all the StackStorm components. The current StackStorm use case involves sensors, events, and triggers that automate the processing of raw genomics data through a complex workflow that is quite large and long-running—it generally finishes within 24 hours. When the last reports are generated on the remote super-computing center, there are manual processes that kick off delivery of the data to the researcher, and in the case of human DNA, also runs a separate downstream best practice pipeline called Piper. Incorporating these downstream processes within the main Mistral workflow, so that more of their work gets automated, is work in progress at the moment.
Besides the workflows for sequence processing, the team is also making heavy usage of Stackstorm traces. They tag all their workflow runs with tags unique for each sequencing run, so that they can go back in history and check all associated executions with their custom script .This comes in handy when troubleshooting and also for auditing purposes (which is important for them as they are an accredited facility).
StackStorm is a powerful event-driven automation platform, which provides the flexibility and autonomy needed for the team to unleash the team’s creativity–providing the freedom to innovate and automate genomic sequencing operations. The team’s work caters to the entire Swedish research community—so serves a very wide variety of research including—Cancer, Cardiovascular disease, and Microbial genetics. A list of publications which have used the lab’s resources is available here.
Two examples of recent interesting pre-prints/publications that have carried out sequencing at their facility are
SweGen: A whole-genome map of genetic variability in a cross section of the Swedish population In which a 1000 Swedish individuals have been sequenced in order to provide a genetic base-line for the population, something which is of great interest to both population genetics, but also in clinical research applications where these samples can then be used as controls.
Complex archaea that bridge the gap between prokaryotes and eukaryotes: A group using the resources were able to identify what is possible the missing link between prokaryotes (bacteria) and eukaryotes (that part of the evolutionary tree to which e.g. humans belong) by sequencing samples from an hydrothermal vent called Loki’s Castle.
As a member of the StackStorm team, it is gratifying to know that StackStorm technology is part of the effort to solve these challenges, and to think of the benefits that this research will bring to millions of people around the world.
Interested to learn how StackStorm can help you address your Genomics or other Big Data challenges? Install StackStorm, join conversation, and help drive innovation at the speed of community.